266
andrias, 19
(2012)
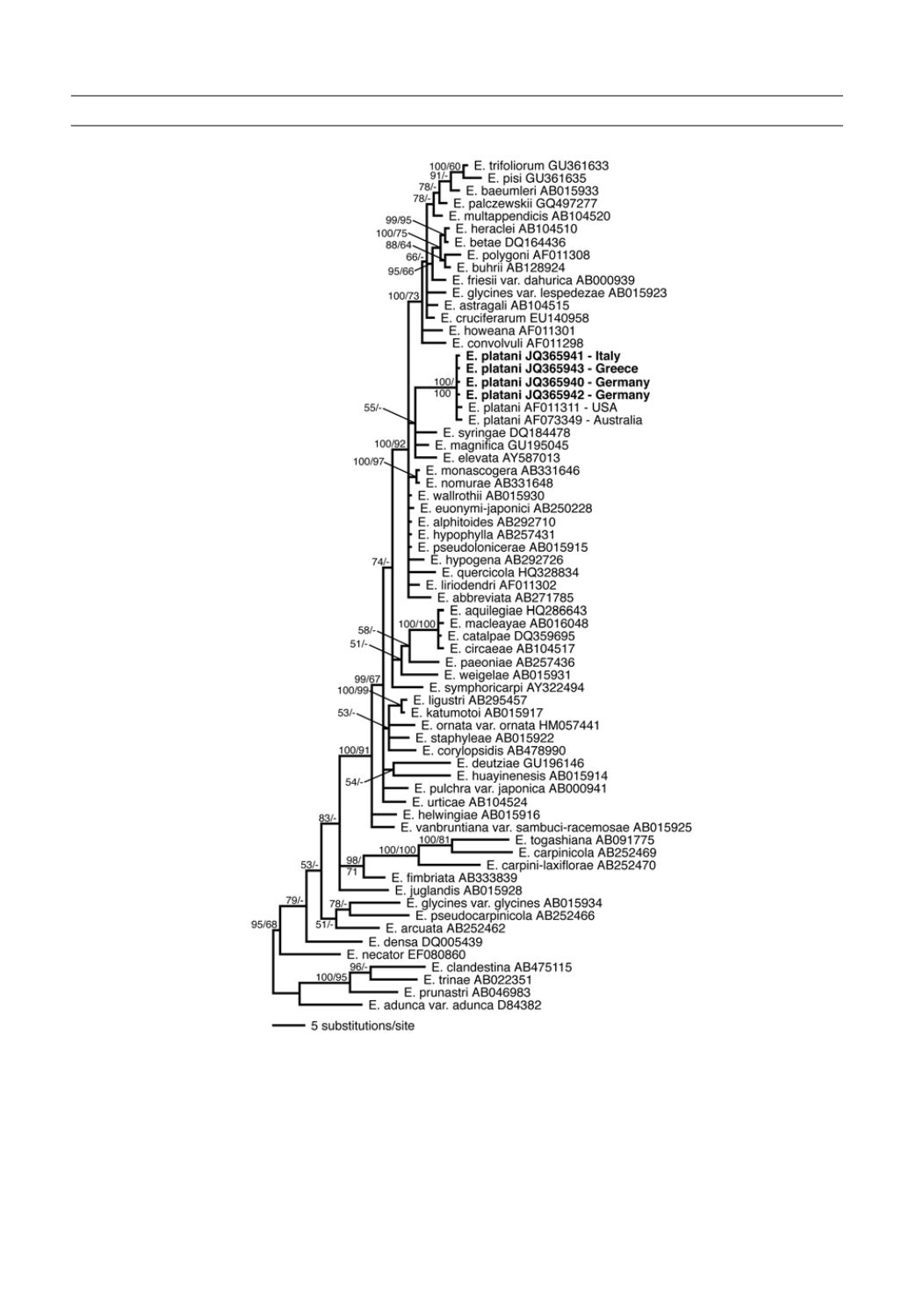
Figure 2.
Bayesian inference of phylogenetic relationships within the sampled Erysiphe species: Markov chain
Monte Carlo analysis of an alignment of ITS base sequences using the GTR+I+G model of DNA substitution with
gamma-distributed substitution rates and estimation of invariant sites, random starting trees and default starting
parameters of the DNA substitution model. A 50% majority-rule consensus tree is shown computed from 45.000
trees that were sampled after the process had reached stationarity. The topology was rooted with Erysiphe adunca
var. adunca, E. clandestina, E. prunastri, and E. trinae. Numbers on branches before slashes are estimates for
a posteriori probabilities, numbers after slashes are ML bootstrap support values. Branch lengths were averaged
over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. Speci-
mens examined in this study are printed in bold. E. = Erysiphe.


